O1 - Typical of Austronesians, southern Han Chinese
O2a - Typical of Austro-Asiatic peoples, Kradai peoples, Malays, Indonesians, and Malagasy,
O2b - origins in Manchuria, followed by increase in Korea. Typical of Koreans/Japan
O3 - Typical of populations of EAsia, SAsia, Austronesian,CentralAsia
References
1. ^ Laura Scheinfeldt, Françoise Friedlaender, Jonathan Friedlaender, Krista Latham, George Koki, Tatyana Karafet, Michael Hammer and Joseph Lorenz, "Unexpected NRY Chromosome Variation in Northern Island Melanesia," Molecular Biology and Evolution 2006 23(8):1628-1641
2. ^ Yali Xue, Tatiana Zerjal, Weidong Bao, Suling Zhu, Qunfang Shu, Jiujin Xu, Ruofu Du, Songbin Fu, Pu Li, Matthew E. Hurles, Huanming Yang, and Chris Tyler-Smith, "Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times," Genetics 2006 April; 172(4): 2431–2439.
3. ^ Supplementary Table 2: NRY haplogroup distribution in Han populations, from the online supplementary material for the article by Bo Wen et al., "Genetic evidence supports demic diffusion of Han culture," Nature 431, 302-305 (16 September 2004)
4. ^ Table 1: Y-chromosome haplotype frequencies in 49 Eurasian populations, listed according to geographic region, from the article by R. Spencer Wells et al., "The Eurasian Heartland: A continental perspective on Y-chromosome diversity," Proceedings of the National Academy of Sciences of the United States of America (August 28, 2001)
5. ^ "Y-Chromosome Evidence for Differing Ancient Demographic Histories in the Americas," Maria-Catira Bortolini et al., American Journal of Human Genetics 73:524-539, 2003
------------------------------------------------------------------------
O2b--> belonging to a man who already resides within Greater Manchuria, or who at least already belongs to a specific "proto-Tungus-Korean" tribe.
http://en.wikipedia.org/wiki/Haplogroup_O2_(Y-DNA)
Although Haplogroup O2 is not found so frequently in modern human populations as its brother clade, Haplogroup O3, it is notable for the peculiarities of its geographical distribution. Like all clades of Haplogroup O, Haplogroup O2 is found only among the males of modern Eastern Eurasian populations. However, unlike Haplogroup O3, which is shared in common by almost all populations of Eastern Eurasia as well as many populations of Oceania, Haplogroup O2 is generally found only among certain populations, such as the Austroasiatic peoples of India, Bangladesh and Southeast Asia, the Nicobarese of the Nicobar Islands in the Indian Ocean, and the Koreans, Japanese, and Tungusic peoples of Northeast Asia.
Besides its widespread and patchy distribution, Haplogroup O2 is also notable for the fact that it can be divided into two major subclades that show almost completely disjoint distribution. One of these subclades, Haplogroup O2a (M95), is found among some (mostly tribal) populations of South and Southeast Asia, as well as among the Khmers of Cambodia and the Balinese of Indonesia. There are also some reports that Haplogroup O2a may be associated with the so-called Negrito populations of mainland Southeast Asia, such as the Semang and the Senoi, but it is not clear whether this is their original Y-chromosome heritage or rather the result of incursion of Austroasiatic Y-chromosomes from the Khmers and related peoples who may have lent the Negritos their Austroasiatic languages in a manner similar to the Bantus' hypothesized lending of their languages and Haplogroup E Y-chromosomes to the pygmoid peoples of Africa, such as the Baka and Mbuti. The other major subclade, Haplogroup O2b (SRY465, M176), is found almost exclusively among the Koreans, the Japanese, and the Manchus.
The Northeast Asian Haplogroup O2b suggests a very interesting sort of relationship between the Tungusic, Korean, and Japanese peoples. Haplogroup O2b's parent haplogroup, Haplogroup O2*, appears to have been thinly spread throughout East Asia since prehistoric times. The derived Haplogroup O2b* (M176) is found at low frequency across the populations of the whole of Greater Manchuria, including the Tungusic, Korean, Japanese, and even the Daur people, who speak a Mongolic language that has traditionally been considered to have derived from an ancient dialect transitional between the Proto-Mongolic and Proto-Tungusic languages. One major subbranch, O2b1* (P49), is shared between the Korean and Japanese populations (with a significantly higher STR diversity among the Koreans), but its distribution does not reach the Tungusic or Daur peoples to the north. Finally, the most derived subbranch, Haplogroup O2b1a (47z), is essentially restricted to the Japanese population, although it has sporadically been detected in a few individuals who reside within the extent of the erstwhile Great Japanese Empire, in which cases it is believed to represent minor admixture from male Japanese soldiers or civilians into the local populations within the last several generations; patrilineal descent from a Japanese visitor or immigrant to some of these lands during earlier historical times is another possibility. This "nested" hierarchical and region-specific distributional pattern tends to suggest the following sequence of events:
1) The M176 mutation that defines the O2b Y-chromosome lineage first arises on an ancestral O2* chromosome belonging to a man who already resides within Greater Manchuria, or who at least already belongs to a specific "proto-Tungus-Korean" tribe.
2) Sometime after the proto-Koreans have branched off from the other populations of the Greater Manchurian region, the P49 mutation that defines Haplogroup O2b1* arises in a proto-Korean male, whose lineage becomes very successful and increases in number among the prehistoric proto-Korean population.
3) One regional subgroup of the proto-Koreans distinguishes itself from the others either culturally or by physically migrating away from their proto-Korean brethren. After the split from the greater proto-Korean population is complete, the 47z mutation that defines Haplogroup O2b1a occurs in a male of this now proto-Japanese population, who produces many male descendants and increases the frequency of the O2b1a lineage among the proto-Japanese. This very specific Haplogroup O2b1a lineage now occupies about 42.5% of the total of Haplogroup O lineages, or about 22.0% of all Y-chromosomes, among the modern Japanese, which probably reflects extreme hegemony of a certain close-knit clan in the history of the Japanese people.
In addition to suggesting a phylogenetic relationship between at least a subset of the ancestors of the Tungusic, Korean, and Japanese peoples, the distribution of Haplogroup O2b and its subbranches also argues strongly against any significant intermingling of any of these peoples during historical times.
The other major Northeast Asian Y-chromosome haplogroup, Haplogroup C3, is also found among the Tungusic, Korean, and Japanese peoples, as well as among the Mongolian peoples, Turkic peoples, and even the Ainu of Japan and the Na-Dené peoples of North America, but Haplogroup C3 displays a cline of frequency opposite to that of Haplogroup O2, to the effect that Haplogroup C3 is more frequent among the more northerly populations (Mongolian, Turkic, Northern Tungusic, Koryak, Nivkh, etc.), whereas Haplogroup O2 is more frequent among the more southerly populations (Southern Tungusic, Korean, and Japanese), and it is entirely absent from the Ainu and Na-Dené populations. The typically Sino-Tibetan and Southeast Asian Haplogroup O3 is also shared in common by most Tungusic, Mongolic, Turkic, Korean, and Japanese populations with various frequency and diversity values, which may be a genetic trace of a demic diffusion of Neolithic culture from China; however, the common occurrence at a low frequency throughout Northeast Asia of Y-chromosome Haplogroup D1 and several mtDNA haplogroups that are typical of Tibetans in addition to the Sino-Tibetan-Austronesian Haplogroup O3 may reflect some sort of relationship between the peoples of Northeast Asia and the peoples of Tibet rather than a major Neolithic or even more recent influence from China.
------------------------------------------------------------------------
Haplogroup O1
http://en.wikipedia.org/wiki/Haplogroup_O1_(Y-DNA)Both Haplogroup O1 and Haplogroup O3 are prototypical Chinese patrilines, and they are believed to have first evolved sometime during the late Pleistocene (Upper Paleolithic) in Southeast Asia. Both O1 and O3 are commonly found among most modern populations of China and Southeast Asia. These two lineages are also presumed to be markers of the Austronesian expansion, probably originating from prehistoric Taiwan or the neighboring southeastern coast of China, when they are found among modern populations of Maritime Southeast Asia or Oceania.[1]
By far the strongest positive correlation between Haplogroup O1 and ethnolinguistic affiliation is that which is observed between this haplogroup and the Austronesians. It is interesting that the peak frequency of Haplogroup O1 is found among the aborigines of Taiwan, precisely the region from which linguists have hypothesized that the Austronesian language family originated. A slightly weaker correlation is observed between Haplogroup O1 and the Han Chinese populations of southern China, as well as between this haplogroup and the Kradai-speaking populations of southern China and Southeast Asia. The distribution of Daic languages in Thailand and other parts of Southeast Asia outside of China has long been believed, for reasons of traditional linguistic geography, to reflect a recent invasion of Southeast Asia by Daic-speaking populations originating from southeastern China, and the somewhat elevated frequency of Haplogroup O1 among the Daic populations, coupled with a high frequency of Haplogroup O2a, which is a genetic characteristic of the Austro-Asiatic peoples of Southeast Asia, suggests that the genetic signature of the Daic peoples' affinity with populations of southeastern China has been weakened due to extensive assimilation of the earlier Austro-Asiatic residents of the lands which the Daic peoples invaded. Also, it has been noted that Haplogroup O1 lineages among populations of continental Southeast Asia outside of China display a reduced level of diversity when compared with populations of South China and insular Southeast Asia, which may be evidence of a bottleneck associated with the westward migration and settlement of ancestral Daic-speaking populations in Indochina.
[edit] Distribution
Haplogroup O1 is generally found wherever its brother haplogroup, Haplogroup O3, is found, although at a frequency much lower than that of Haplogroup O3. A conspicuous exception to this general pattern is presented by the Taiwanese aboriginal populations, among whom Haplogroup O1 generally dominates Haplogroup O3 in frequency. The frequency of Haplogroup O1 has been found to be negatively correlated with latitude, so that, on average, it occurs at relatively higher frequency towards the more southerly parts of its range, but it never attains majority haplogroup status outside of Taiwan. Heightened frequencies of Haplogroup O1 have also been observed in samples of populations from the Philippines and the eastern-southeastern coast of China. This haplogroup is, however, conspicuously absent from populations of the Japanese Archipelago, which seems to preclude a close genetic relationship between the peoples of Japan and the Austronesians, despite many enthusiastic attempts to prove a common origin of the Japanese and Austronesian languages. Haplogroup O1a-M119 Y-chromosomes have also been found to occur at low frequency among various populations of Siberia, such as the Nivkhs (one of 17 sampled Y-chromosomes), Ulchi/Nanai (2/53), Yenisey Evenks (1/31), and especially the Buryats living in the Sayan-Baikal uplands of Irkutsk Oblast (6/13).[2]
[edit] Frequencies
The frequencies of Haplogroup O1 among various East Asian and Austronesian populations suggest a complex genetic history of the modern Han populations of southern China. Although Haplogroup O1 occurs only at an average frequency of approximately 4% among Han populations of northern China and peoples of southwestern China and Southeast Asia who speak Tibeto-Burman languages, the frequency of this haplogroup among the Han populations of southern China nearly quadruples to about 15%. It is particularly interesting that the frequency of Haplogroup O1 among the Southern Han has been found to be slightly greater than the arithmetic mean of the frequencies of Haplogroup O1 among the Northern Han and a pooled sample of Austronesian populations. This suggests that modern Southern Han populations possess a non-trivial number of male ancestors who were originally affiliated with some Austronesian-related culture, or who at least shared a genetic affinity with many of the ancestors of modern Austronesian peoples.
------------------------------------------------------------------------
Haplogroup O3 (Y-DNA)
http://en.wikipedia.org/wiki/Haplogroup_O3_(Y-DNA)Origins
Origins
Haplogroup O3 is a descendant haplogroup of haplogroup O. Some researchers believe that it first appeared in China approximately 10,000 years ago. However, others believe that the high internal diversity of Haplogroup O3 indicates a Late Pleistocene (Upper Paleolithic) origin in South China or Southeast Asia of the M122 mutation that defines the entire O3 clade, while the common presence among a wide variety of modern East and Southeast Asian nations of closely related haplotypes belonging to certain subclades of Haplogroup O3 is considered to point to a recent (e.g., Holocene) geographic dispersion of a certain subset of the ancient variation within Haplogroup O3. The spread of these particular subsets of Haplogroup O3 is conjectured to be closely associated with the sudden agricultural boom associated with rice farming.
The prehistoric peopling of East Asia by modern humans remains controversial with respect to early population migrations. In a systematic sampling and genetic screening of an East Asian–specific Y-chromosome haplogroup (O3-M122) in 2,332 individuals from diverse East Asian populations. Results indicate that the O3-M122 lineage is dominant in East Asian populations, with an average frequency of 44.3%. The microsatellite data show that the O3-M122 haplotypes in southern East Asia are more diverse than those in northern East Asia, suggesting a southern origin of the O3-M122 mutation. It was estimated that the early northward migration of the O3-M122 lineages in East Asia occurred ~25,000–30,000 years ago, consistent with the fossil records of modern humans in East Asia. [1]
[edit] Distribution
Although Haplogroup O3 appears to be primarily associated with Chinese populations, it also forms a significant component of the Y-chromosome diversity of most modern populations of the East Asian region. Haplogroup O3 is found in over 50% of all modern Chinese males (ranging up to over 80% in certain regional subgroups of the Han ethnicity), about 40% of Manchurian, Korean, and Vietnamese males, about 33.3%[2] to 60.7%[3] of Filipino males, about 35% of Malaysian males, about 25% of Zhuang[4] and Indonesian[5] males, and about 15%[6] to 20%[2] of Japanese males. The distribution of Haplogroup O3 stretches far into Central Asia (approx. 30% of Salar, 24% of Dongxiang[7], 18% to 22.8% of Mongolians, 12% of Uyghurs, 9% of Kazakhs, 6.2% of Altayans[8], and 4.1% of Uzbeks) and Oceania (approx. 25% of Polynesians, 18% of Micronesians, and 5% of Melanesians[9]), albeit with reduced frequencies of most subclades. It should be noted that Haplogroup O3* Y-chromosomes, which are not defined by any identified downstream markers, are actually more common among certain non-Han Chinese populations than among Han Chinese ones, and the presence of these O3* Y-chromosomes among various populations of Central Asia, East Asia, and Oceania is more likely to reflect a very ancient shared ancestry of these populations rather than the result of any historical events. It remains to be seen whether Haplogroup O3* Y-chromosomes can be parsed into distinct subclades that display significant geographical or ethnic correlations.
Among all the populations of East and Southeast Asia, Haplogroup O3 is most closely associated with those that speak a Sinitic, Tibeto-Burman, or Hmong-Mien language. Haplogroup O3 comprises about 50% or more of the total Y-chromosome variation among the populations of each of these language families. The Sinitic and Tibeto-Burman language families are generally believed to be derived from a common Sino-Tibetan protolanguage, and most linguists place the homeland of the Sino-Tibetan language family somewhere in northern China. The Hmong-Mien languages and cultures, for various archaeological and ethnohistorical reasons, are also generally believed to have derived from a source somewhere north of their current distribution, perhaps in northern or central China. The Tibetans, however, despite the fact that they speak a language of the Tibeto-Burman language family, have high percentages of the otherwise rare haplogroups D1 and D3, which are also found at much lower frequencies among the members of some other ethnic groups in East Asia and Central Asia.
Haplogroup O3 has been implicated as a diagnostic genetic marker of the Austronesian expansion when it is found in populations of Oceania. Its distribution in Oceania is mostly limited to the traditionally Austronesian culture zones, including moderately high frequencies in the Philippines, Malaysia, Indonesia, and Polynesia, with generally lower frequencies found in coastal and island Melanesia, Micronesia, and Taiwanese aboriginal tribes.
The subgroup O3a5-M134 is particularly closely associated with Sino-Tibetan populations, and it is generally not found outside of areas where a Sino-Tibetan language is currently spoken or that are historically supposed to have undergone Chinese colonization or immigration, such as Korea, Japan, Vietnam, Malaysia, the Philippines, and Indonesia. However, its presence among non-Sino-Tibetan populations is always very limited and never amounts to more than 10% of the total Y-chromosome diversity. There are also reports that Y-chromosomes belonging to Haplogroup O3a5 have been sampled from populations of such far-flung places as Western Samoa. Surprisingly, Haplogroup O3a5-M134 Y-chromosomes have also been found in about 1% to 3% of indigenous Australian men in the northwest of that continent, which might indicate that a certain degree of contact has occurred between the Austronesian expansion from Asia and some indigenous Australian populations. The fact that Haplogroup O3a5 is so strongly associated with Chinese populations, however, and the fact that no Y-chromosome haplogroups characteristic of Austronesian populations have been found among these indigenous Australian populations may be taken to suggest the possibility of some direct Chinese-Australian contact in the precolonial era. Within Japan, the subgroup O3a5-M134 forms the majority of the haplogroup O3 Y-chromosomes detected.
Haplogroup O3's brother clade, Haplogroup O1, displays a similar geographical distribution, being found among nearly all the populations of East and Southeast Asia, but generally at a frequency much lower than that of Haplogroup O3. Another brother clade, Haplogroup O2, has an impressive extent of dispersal, as it is found among the males of populations as widely separated as the Kolarians of India and the Japanese of Japan; however, Haplogroup O2's distribution is much more patchy, and the Haplogroup O2 Y-chromosomes found among the Mundas and the Japanese belong to distinct subclades.
[edit] References
1. ^ Hong Shi, Yong-li Dong, Bo Wen, Chun-Jie Xiao, Peter A. Underhill, Pei-dong Shen, Ranajit Chakraborty, Li Jin, and Bing Su, "Y-Chromosome Evidence of Southern Origin of the East Asian–Specific Haplogroup O3-M122" American Journal of Human Genetics 77:408–419, 2005 (HTML) (PDF)
2. ^ a b Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes, Michael F. Hammer et al., Journal of Human Genetics (Jan. 2006)
3. ^ Matthew E. Hurles, Bryan C. Sykes, Mark A. Jobling, and Peter Forster, "The Dual Origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from Maternal and Paternal Lineages," American Journal of Human Genetics 76:894–901, 2005.
4. ^ Chen Jing, Li Hui et al., "Y-chromosome Genotyping and Genetic Structure of Zhuang Populations," Acta Genetica Sinica (Dec. 2006)
5. ^ Hui Li, Bo Wen, Shu-Juo Chen, Bing Su, Patcharin Pramoonjago, Yangfan Liu, Shangling Pan, Zhendong Qin, Wenhong Liu, Xu Cheng, Ningning Yang, Xin Li, Dinhbinh Tran, Daru Lu, Mu-Tsu Hsu, Ranjan Deka, Sangkot Marzuki, Chia-Chen Tan and Li Jin, "Paternal genetic affinity between western Austronesians and Daic populations," BMC Evolutionary Biology 2008, 8:146 doi:10.1186/1471-2148-8-146. http://www.biomedcentral.com/1471-2148/8/146
6. ^ Y-chromosomal Binary Haplogroups in the Japanese Population and their Relationship to 16 Y-STR Polymorphisms, I. Nonaka et al., Annals of Human Genetics (Feb. 2007)
7. ^ Wei Wang, Cheryl Wise, Tom Baric, Michael L. Black and Alan H. Bittles, "The origins and genetic structure of three co-resident Chinese Muslim populations: the Salar, Bo'an and Dongxiang," Human Genetics (2003)
8. ^ V. N. Kharkov, V. A. Stepanov, O. F. Medvedeva, M. G. Spiridonova, M. I. Voevoda, V. N. Tadinova and V. P. Puzyrev, "Gene pool differences between Northern and Southern Altaians inferred from the data on Y-chromosomal haplogroups," Russian Journal of Genetics, Volume 43, Number 5 (May, 2007)
9. ^ Balinese Y-Chromosome Perspective on the Peopling of Indonesia: Genetic Contributions from Pre-Neolithic Hunter-Gatherers, Austronesian Farmers, and Indian Traders, Tatiana M. Karafet, J. S. Lansing, Alan J. Redd, Joseph C. Watkins, S. P. K. Surata, W. A. Arthawiguna, Laura Mayer, Michael Bamshad, Lynn B. Jorde, and Michael F. Hammer, Human Biology (Feb. 2005)
9. Gan Rui-Jing, Pan Shang-Ling, Mustavich Laura F, Qin Zhen-Dong, Cai Xiao-Yun, Qian Ji, Liu Cheng-Wu, Peng Jun-Hua, Li Shi-Lin, Xu Jie-Shun, Jin Li, Li Hui*: the Genographic Consortium (2008) Pinghua Population as an Exception of Han Chinese's Coherent Genetic Structure [1]. J Hum Genet 53:303-313.

here you go.
Koreans
C3- 17%
N3- 4%
N- 8%
O2b- 35%
O3- 30%
Japanese
C3- 5% (C3a)
N3- none
N- <1%
O2b- 35%
O3- 15%
Northern Han
C3- 10% (C3d)
N3- none
N- 7%
O2b- none
O3- 55%
Manchu
C3- 17%
N3- none
N- 2%
O2b- 34%
O3- 43%
http://www.imbice.org.ar/es/lab_06_b/06.pdf
-------------------------------------------------------------------------
http://images.google.co.kr/imgres?imgurl=http://www.mediamob.co.kr/FDS/newBlogContent/2006/0419/imwons/%EA%B3%A0%EB%8C%80%ED%95%9C%EA%B5%AD%EB%AC%B8%EB%AA%854.gif&imgrefurl=http://www.mediamob.co.kr/imwons/frmView.aspx%3Fid%3D85335&usg=__3Ymkf0JlmHvn_dJo27X41SR6de8=&h=529&w=670&sz=28&hl=ko&start=1&um=1&tbnid=AAk5iAKrorCYmM:&tbnh=109&tbnw=138&prev=/images%3Fq%3DFarmers%2Band%2BTheir%2BLanguages:%2BThe%2BFirst%2BExpansions%26hl%3Dko%26client%3Dfirefox-a%26rls%3Dorg.mozilla:en-US:official%26sa%3DN%26um%3D1
http://pritch.bsd.uchicago.edu/publications/FalushEtAl03_Science.pdf
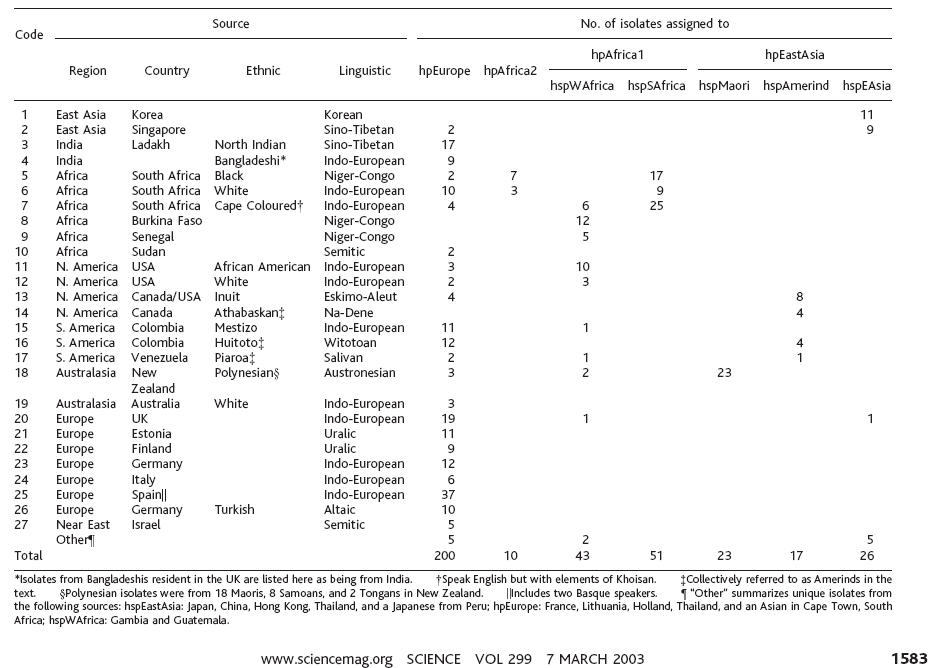
"Traces of Human Migrations in Helicobacter pylori Populations"
Daniel Falush, Thierry Wirth, Bodo Linz, Jonathan K. Pritchard, Matthew Stephens, Mark Kidd, Martin J. Blaser, David Y. Graham, Sylvie Vacher, Guillermo I. Perez-Perez, Yoshio Yamaoka, Francis Mégraud, Kristina Otto, Ulrike Reichard, Elena Katzowitsch, Xiaoyan Wang, Mark Achtman, and Sebastian Suerbaum
Science 7 March 2003; 299: 1582-1585
Daniel Falush, Thierry Wirth, Bodo Linz, Jonathan K. Pritchard, Matthew Stephens, Mark Kidd, Martin J. Blaser, David Y. Graham, Sylvie Vacher, Guillermo I. Perez-Perez, Yoshio Yamaoka, Francis Mégraud, Kristina Otto, Ulrike Reichard, Elena Katzowitsch, Xiaoyan Wang, Mark Achtman, and Sebastian Suerbaum
Science 7 March 2003; 299: 1582-1585
"Farmers and Their Languages: The First Expansions "
Jared Diamond and Peter Bellwood
Science 25 April 2003; 300: 597-603
Jared Diamond and Peter Bellwood
Science 25 April 2003; 300: 597-603



-------------------------------------------
According to the latest 2007 genetic/DNA study by Jiang Ya-ping at Labs of Cellular and Molecular Evolution in Kunming Institute of Zoology, China,
the , contradicting the theory of "Beijing Apes"
No comments:
Post a Comment